PyTomography#
PyTomography is a python library for medical image reconstruction. It uses the functionality of PyTorch to (i) enable fast GPU-accelerated reconstruction and (ii) permit easy integration of deep-learning models in traditional reconstruction algorithms. If you use PyTomography in your own research, please cite the following: https://arxiv.org/abs/2309.01977
Features#
Modalities
Single Photon Computed Emission Tomography (SPECT)
Support for attenuation correction, PSF modeling, and energy window based scatter correction
Support for SIMIND and DICOM data, including stitching of multiple bed positions
Positron Emission Tomography (PET)
Support for sinogram/listmode and time-of-flight reconstruction with attenuation, normalization, random, and scatter correction (time-of-flight based single scatter simulation)
Support for GATE, HDF5, and PETSIRD data formats
Reconstruction Algorithms
Installation#
Firstly, it is important that all your graphics card drivers are up to date. To ensure this, go to this link, enter your graphics card information, and download/install the newest possible driver. It is also recommended that you download anaconda using this link to manage your python environments.
Firstly, create a pytomography virtual environment using anaconda; the library requires python 3.11 or higher:
conda create --name pytomography_env -c conda-forge python=3.11
Then you can activate your environment, and install PyTomography:
conda activate pytomography_env
pip install pytomography
If you want to use the PET reconstruction options available, you need to additionally follow the installation instructions for parallelproj at this link.
Tutorials#
Be sure to check out Tutorials for some simple examples. If you wish to make a contribution, please read the Developer’s Guide.
User Forum#
If you have any questions about how to use the software, feel free to ask them on the discourse page at this link.
Examples#
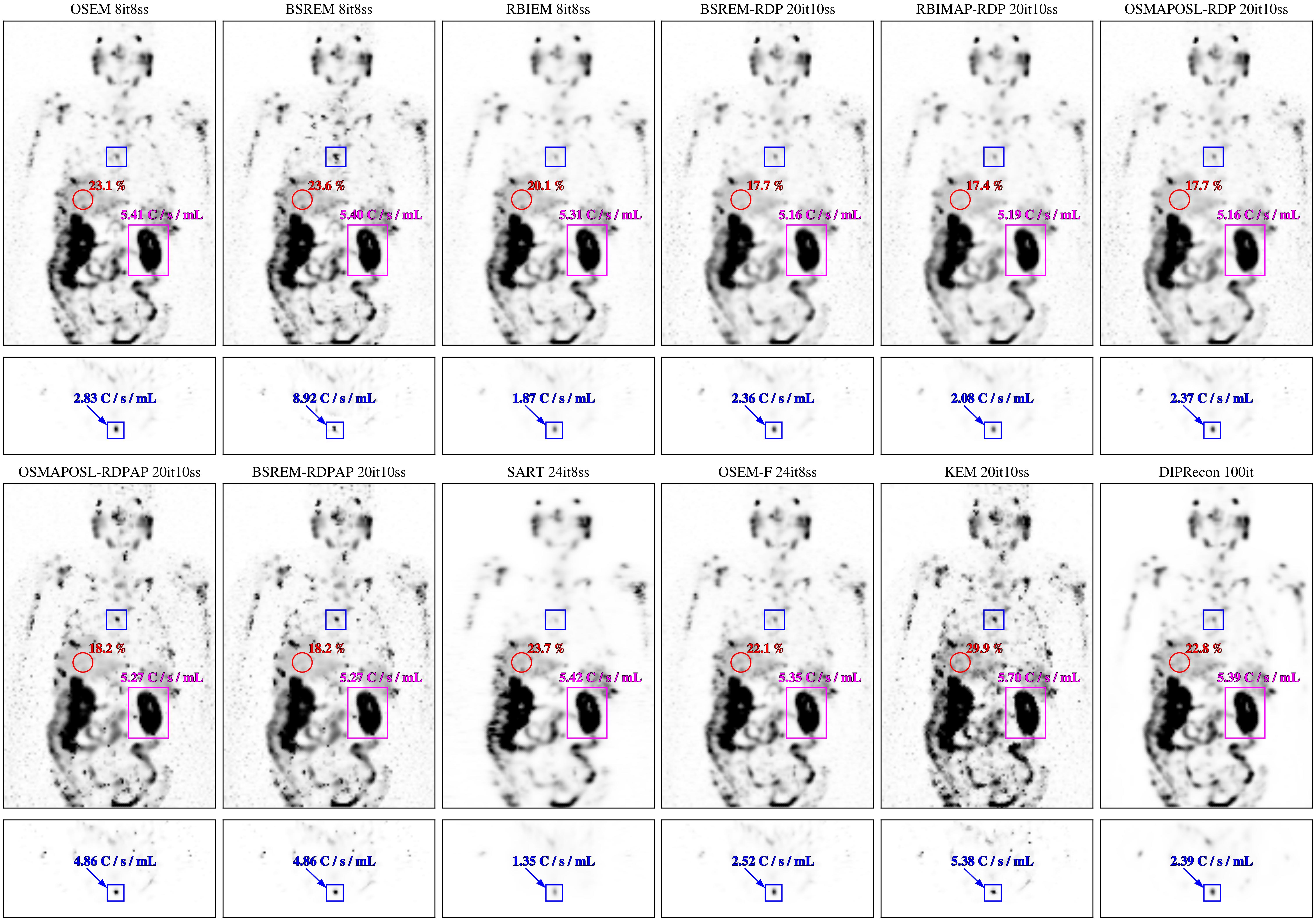
Example 1: Reconstruction of Lu177-PSMA patient data using the available reconstruction algorithms.
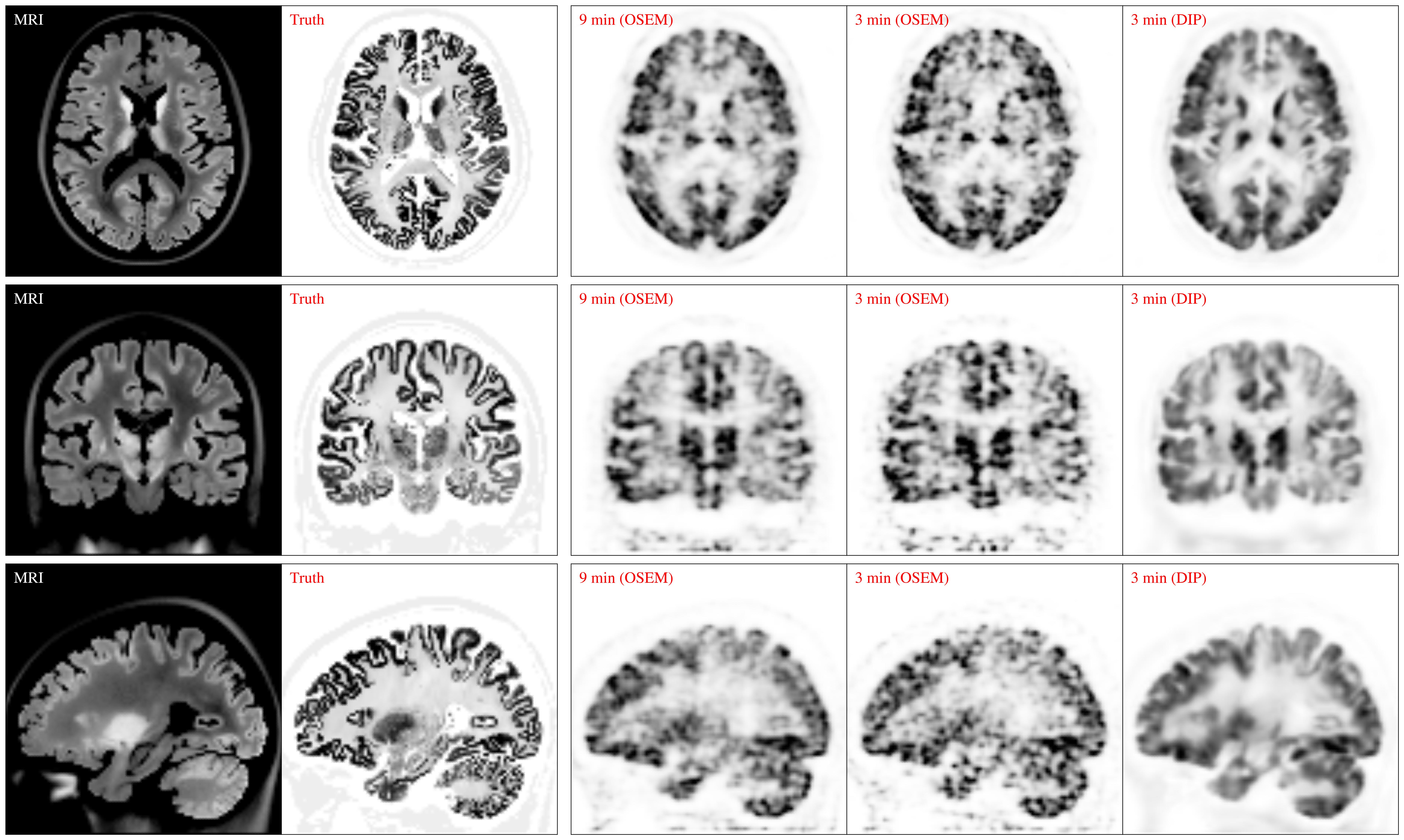
Example 2: Ultra high resolution PET/MR brain phantom reconstructed using list-mode time-of-flight reconstruction with standard OSEM and the AI-Based Deep Image Prior reconstruction algorithm. Scatter estimation (listmode/time-of-flight) was obtained via single scatter simulation (SSS).